A lot of my work is about making tight loops.
Or maybe it’s just that as I gain mastery (slowly) of programming on datasets, the effort I expend is more around the edges of that, and the more I do that the more I understand this is a problem domain in an of itself.
For me, a tight loop is usually made by the scaffolding around the project. Typically this scaffold also made from code like the project. The scaffold’s job is to allow me to observe the effects of the project code that I write rapidly, ideally instantaneously, and to paper over context switching to keep me focussed.
The more rapidly I can get feedback the quicker I can learn if I’m on the right track and correct to eventually complete the task. While context switching and focus are intimately, and invsersely related in my experience.
In R, {targets} is a tool for making tight loops. You change a pipeline, and by the magic of caching, get to observe the effect of that change in the shorest possible time.
Likewise, {rmarkdown} is a tool for making tight loops, but focussed on scientific documents that contain assets built from code. We can change the code that builds the assets, and immediately view the resulting document, without all the slow GUI work of attaching new figures etc.
{testthat} and test suites in general are tools for rapidly collecting vast amounts of feedback about a software project. When adding new code, we can very quickly evaluate if that code has created any problems with existing functionality.
And then there’s the R REPL itself! A spectacular facilitator of tight loops for making graphics, munging data, modelling data etc. I’m supremely spoiled when I use R.
I feel the absence of these loops when they’re missing. The last project I worked on before this extended COVID break was updating a small vector tile server written in Typescript on AWS lambda. I didn’t create the project, and it was initially extremely disorienting trying to get a workflow going.
One thing I knew was that a workflow that was like: Compile to JS, zip code, upload to AWS lambda, test in browser… was not going to work for me. That feels like sludge. I could work like that, but it would cause a lot of bad feelings, and probably take even longer because I’d keep getting distracted between all the context switches.
After some research, I decided to go the “infrastructure as code” route with AWS SAM. By virtue of doing that, I got to run the lambda function locally in a simulated environment, including attaching VSCode’s debugger! That’s pretty tight. I spent probably a week setting that up before even looking into making the changes I needed to make. I think a less experienced me would have felt the pressure to just get in there and start hacking, eager to show progress. But I was able to sell the infrastructure investment to the team with the confidence that it would all be worth it once I had the tight loop rolling.
I see this need of mine cropping up in other places too: I’ve been casually working on my on bespoke keyboard design. Taking what I learned building a Corne, but realising that in a design for my unique and hand measurements.
Initially, I was physically laying out the keyboard in KiCad, and doing paper tests on a printed version. Each time I decided to move a key, I had to do a bunch of trigonometry and sometimes propagate that to many affected keys. It got quite cumbersome.
Then I discovered (well re-discovered) Ergogen (thanks Kyle Mitchell) which is a kind of parameterised framework for generating minimal ergonomic keyboard designs from configuration files. Basically AWS SAM for keyboards if you will.
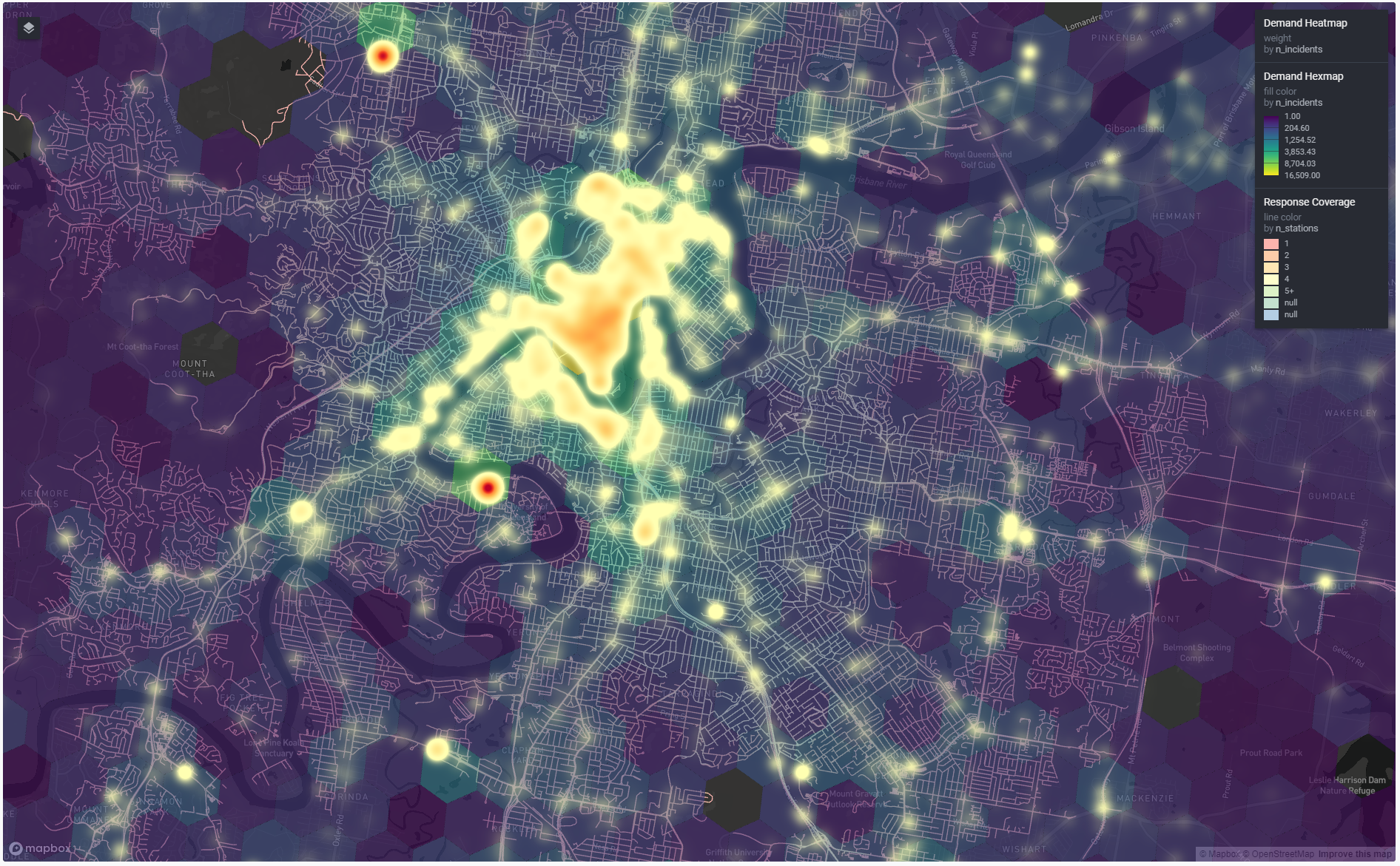
One thing I noticed was that Ergogen doesn’t ship with a quick way to do a complete visualisation of the design. But I was able to close that loop by tacking a little R script on to the build process that read in the outputs as spatial data in {sf} and plotted them with {ggplot2}.
If I have the plot image open in VSCode, it’s automatically refreshed every time I build the config. Tight loop achieved!
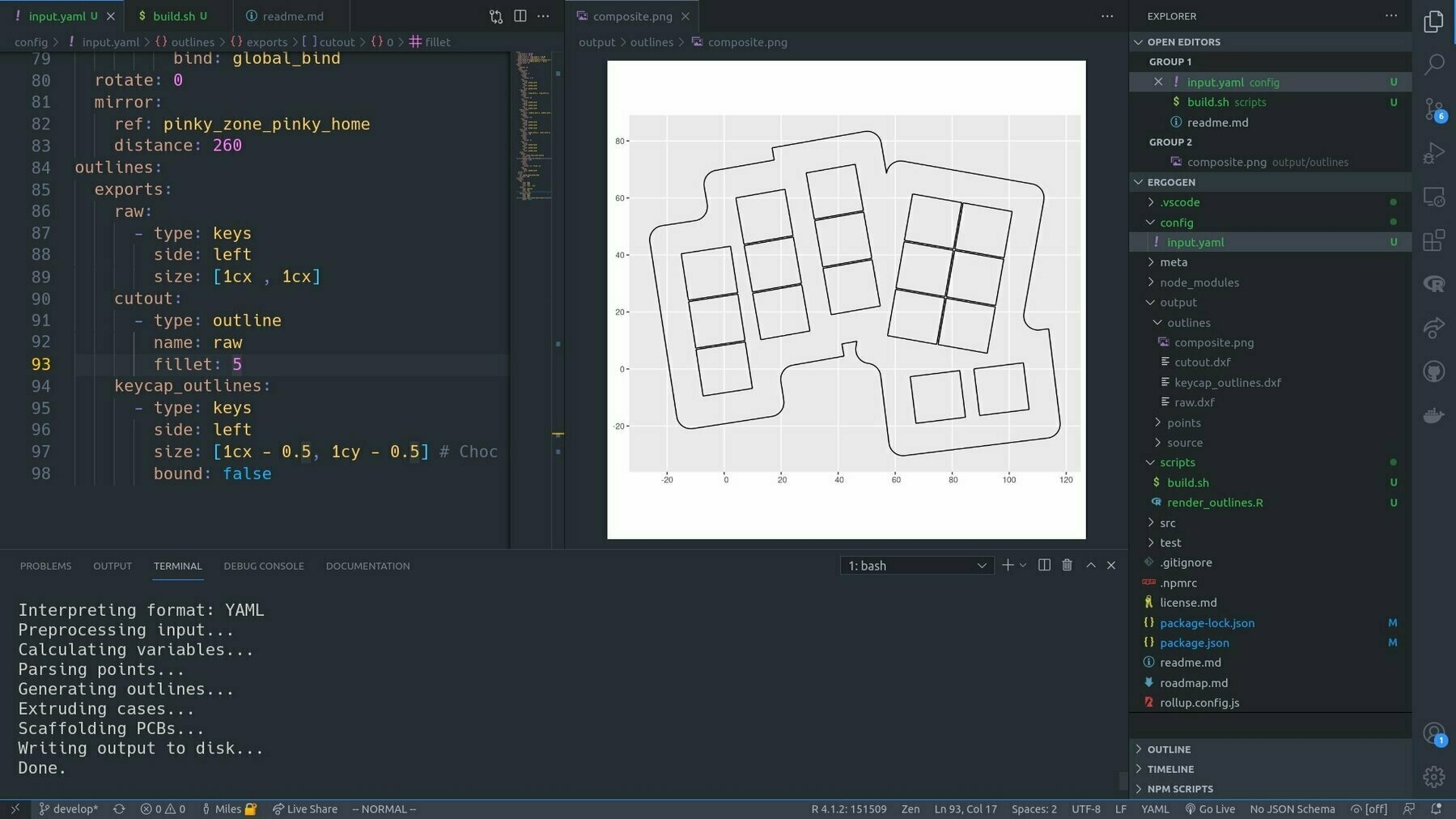
It’s such a powerful concept: investing in the infrastructure of the doing, to boost feedback and minimise context switching. It’s the UI for building the UI. Developer UI. Meta UI?
On reflection, I’ve been into meta UI stuff for a while now. At some level, pretty much all my open source projects are about tightening the loop: Smoothing out annoying snarls that slow down project iteration speed.
Thinking about valuing these things in this context is new for me though.