For #rstats #adventofcode day 2 I decided to avoid all string parsing/manipulation/comparisons and use the command as a class to dispatch s3 methods. Is this a good idea? Probably not!
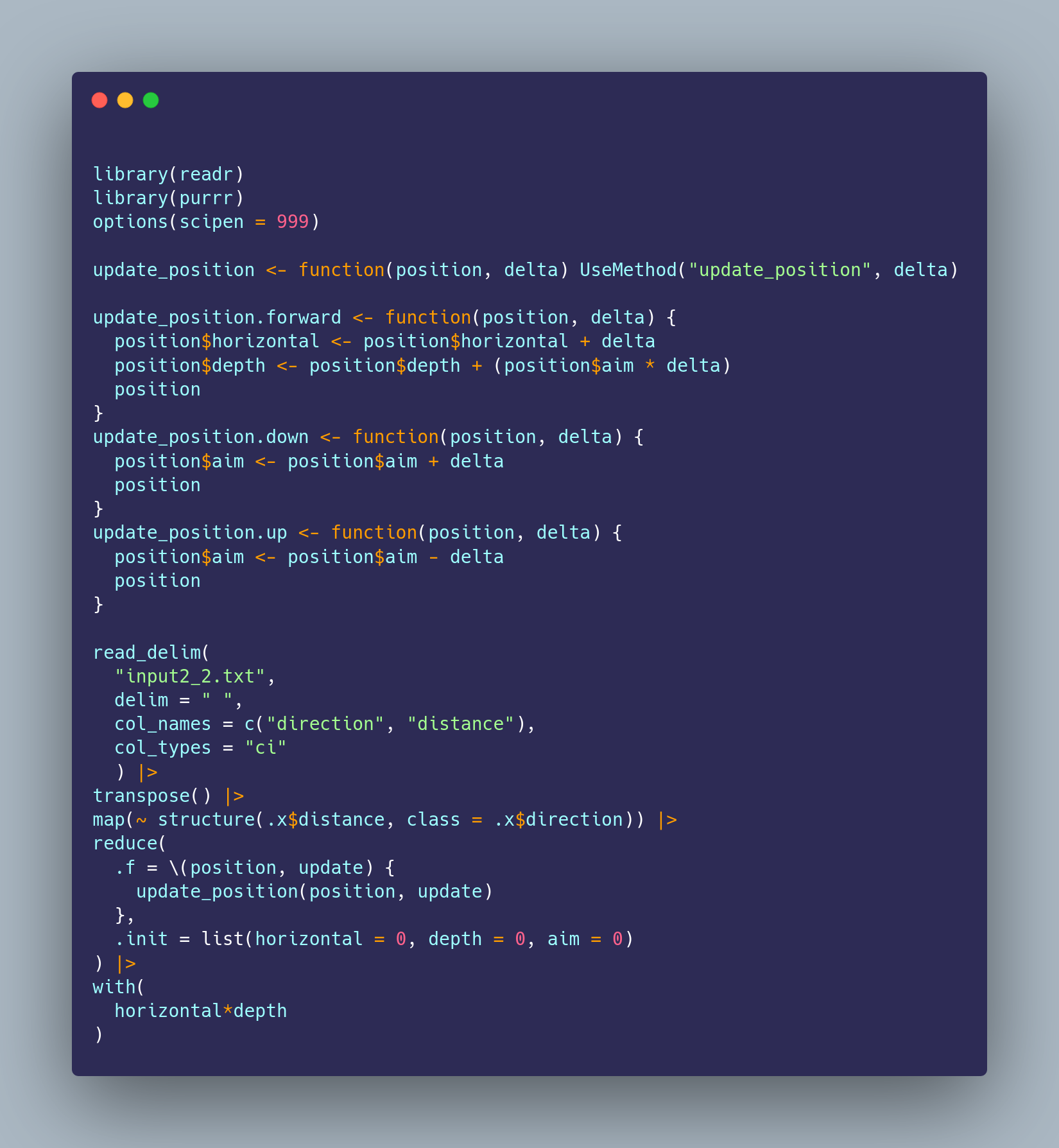
For #rstats #adventofcode day 2 I decided to avoid all string parsing/manipulation/comparisons and use the command as a class to dispatch s3 methods. Is this a good idea? Probably not!

Happy Friday #rstats {targets}/{tflow} users! Added two new addins to help smooth multi-plan workflows: Load target at cursor if found in any store in the _targets.yaml, and tar_make() the active editor plan.
Having a second stab at a plot of my team’s commits since it has come to light that an unnamed someone was using a gmail for user.email on most of their work commits:
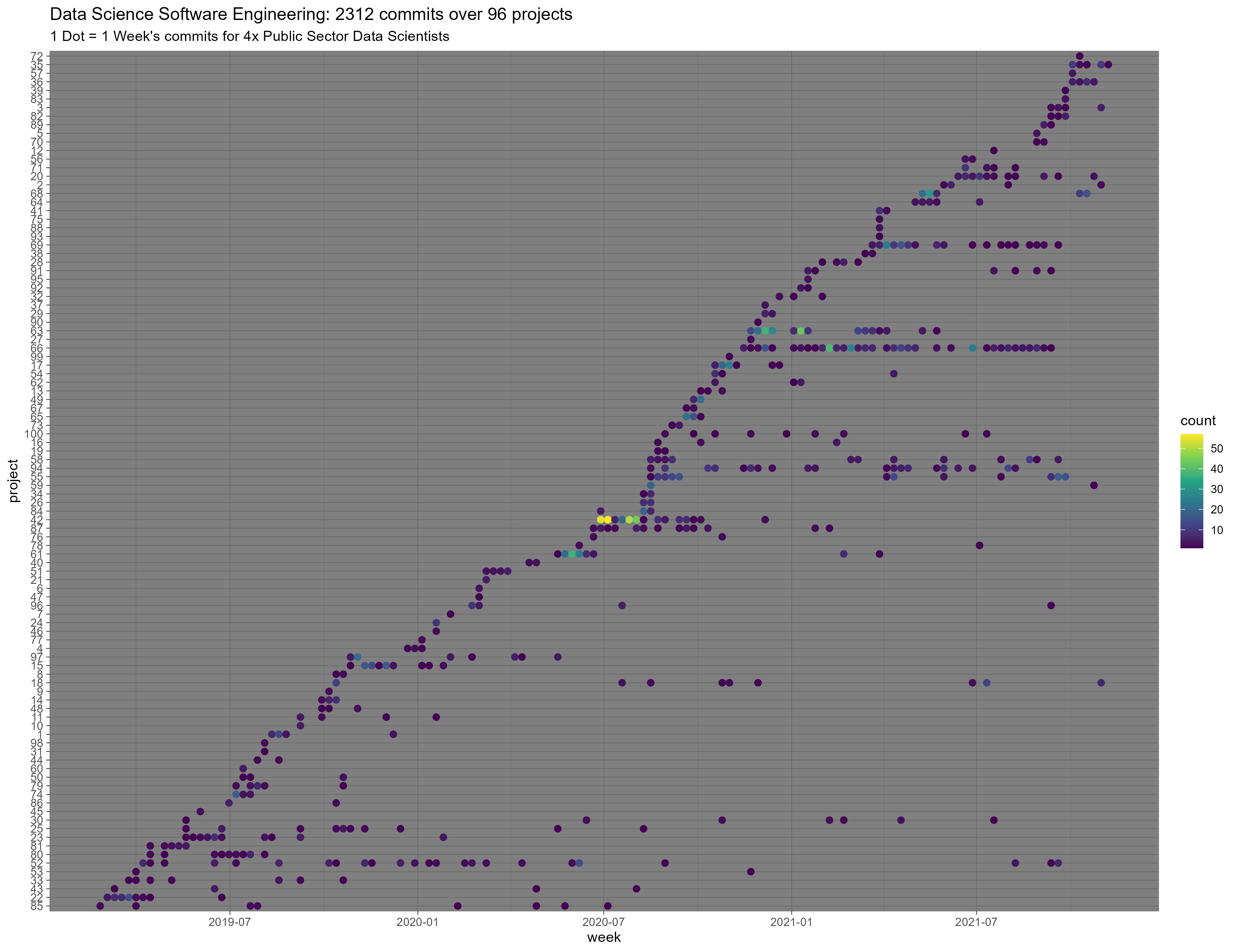
https://cdn.uploads.micro.blog/27894/2021/2c62b05d4a.png
I also binned the dots, instead of using alpha, and of course, that works a lot better.
Regarding software engineering for data science, I think this highlights some important issues I am going to expand on in an upcoming long-form piece. As a teaser: In a world with so many projects, and code constantly flowing between those projects, are “project-oriented workflows” that end their opinions at the project folder underfitting the needs of Data Science teams?
If you’re feeling brave I would love to compare patterns with other teams!
It’s hardly any code (if you have a flat repository structure like mine) thanks to the {gert} package:
library(gert)
library(withr)
library(tidyverse)
library(lubridate)
scan_dir <- "c:/repos"
repos <- list.dirs(scan_dir, recursive = FALSE)
all_commits <- map_dfr(repos, function(repo) {
with_dir(repo, {
branches <- git_branch_list() |> pluck("name")
repo_commits <- map_dfr(branches, function(branch) {
commits <- git_log(ref = branch)
commits$branch <- branch
commits
})
repo_commits$repo <- repo
repo_commits
})
})
qfes_commits <-
all_commits |>
filter(grepl("@qfes|North", author))
duplicates <- duplicated(qfes_commits$commit)
p <-
qfes_commits |>
filter(!duplicates) |>
group_by(repo) |>
mutate(first_commit = min(time)) |>
mutate(repo_num = cur_group_id()) |>
ungroup() |>
group_by(repo_num, first_commit, week = floor_date(time, "week")) |>
summarise(
count = n(),
.groups = "drop"
) |>
ggplot(aes(
x = week,
y = fct_reorder(as.character(repo_num), first_commit),
colour = count
)) +
geom_point(size = 2) +
labs(
title = "Data Science Software Engineering: 2312 commits over 96 projects",
subtitle = "1 Dot = 1 Week's commits for 4x Public Sector Data Scientists",
y = "project"
) +
scale_colour_viridis_c() +
theme_dark()
ggsave(
"commits.png",
p,
device = ragg::agg_png,
height = 10,
width = 13
)
Often when outputting stuff to a package user, the question arises: how much effort could I be bothered to put into formatting the output? The format() function in R has some really nice stuff for this, in particular: alignment.
So today I’m outputting a list of packages to be updated:
arrow 5.0.0.2 -> 6.0.0.2
broom 0.7.7 -> 0.7.9
cachem 1.0.5 -> 1.0.6
cli 3.0.1 -> 3.1.0
crayon 1.4.1 -> 1.4.2
desc 1.3.0 -> 1.4.0
e1071 1.7-8 -> 1.7-9
future 1.22.1 -> 1.23.0
gargle 1.1.0 -> 1.2.0
generics 0.1.0 -> 0.1.1
gert 1.3.2 -> 1.4.1
googledrive 1.0.1 -> 2.0.0
googlesheets4 0.3.0 -> 1.0.0
haven 2.4.1 -> 2.4.3
htmltools 0.5.1.1 -> 0.5.2
jsonvalidate 1.1.0 -> 1.3.1
knitr 1.34 -> 1.36
lattice 0.20-44 -> 0.20-45
lubridate 1.7.10 -> 1.8.0
lwgeom 0.2-7 -> 0.2-8
mime 0.11 -> 0.12
osmdata 0.1.6.007 -> 0.1.8
paws.common 0.3.12 -> 0.3.14
pillar 1.6.3 -> 1.6.4
pkgload 1.2.1 -> 1.2.3
qfesdata 0.2.9011 -> 0.2.9030
reprex 2.0.0 -> 2.0.1
rmarkdown 2.9 -> 2.10
roxygen2 7.1.1 -> 7.1.2
RPostgres 1.3.3 -> 1.4.1
rvest 1.0.1 -> 1.0.2
sf 1.0-2 -> 1.0-3
sodium 1.1 -> 1.2.0
stringi 1.7.4 -> 1.7.5
tarchetypes 0.2.0 -> 0.3.2
targets 0.7.0.9001 -> 0.8.1
tibble 3.1.4 -> 3.1.5
tinytex 0.32 -> 0.33
travelr 0.7.5 -> 0.9.1
tzdb 0.1.2 -> 0.2.0
usethis 2.0.1 -> 2.1.3
xfun 0.24 -> 0.27
Made by this code:
cat(
paste(
lockfile_deps$name,
lockfile_deps$version_lib,
" -> ",
lockfile_deps$version_lock
),
sep = "\n"
)
And one thing that would make it look a bit less amateurish is alignment. I laboured over this sort of stuff years ago when I wrote {datapasta} making really hard work of it - it was the source of an infamous recurring bug. This was partly because I didn’t know that if you call format() on a character vector it automatically pads all your strings to the same length:
e.g.
cat(
paste(
format(lockfile_deps$name),
format(lockfile_deps$version_lib),
" -> ",
format(lockfile_deps$version_lock)
),
sep = "\n"
)
Makes the output look like:
arrow 5.0.0.2 -> 6.0.0.2
broom 0.7.7 -> 0.7.9
cachem 1.0.5 -> 1.0.6
cli 3.0.1 -> 3.1.0
crayon 1.4.1 -> 1.4.2
desc 1.3.0 -> 1.4.0
e1071 1.7-8 -> 1.7-9
future 1.22.1 -> 1.23.0
gargle 1.1.0 -> 1.2.0
generics 0.1.0 -> 0.1.1
gert 1.3.2 -> 1.4.1
googledrive 1.0.1 -> 2.0.0
googlesheets4 0.3.0 -> 1.0.0
haven 2.4.1 -> 2.4.3
htmltools 0.5.1.1 -> 0.5.2
jsonvalidate 1.1.0 -> 1.3.1
knitr 1.34 -> 1.36
lattice 0.20-44 -> 0.20-45
lubridate 1.7.10 -> 1.8.0
lwgeom 0.2-7 -> 0.2-8
mime 0.11 -> 0.12
osmdata 0.1.6.007 -> 0.1.8
paws.common 0.3.12 -> 0.3.14
pillar 1.6.3 -> 1.6.4
pkgload 1.2.1 -> 1.2.3
qfesdata 0.2.9011 -> 0.2.9030
reprex 2.0.0 -> 2.0.1
rmarkdown 2.9 -> 2.10
roxygen2 7.1.1 -> 7.1.2
RPostgres 1.3.3 -> 1.4.1
rvest 1.0.1 -> 1.0.2
sf 1.0-2 -> 1.0-3
sodium 1.1 -> 1.2.0
stringi 1.7.4 -> 1.7.5
tarchetypes 0.2.0 -> 0.3.2
targets 0.7.0.9001 -> 0.8.1
tibble 3.1.4 -> 3.1.5
tinytex 0.32 -> 0.33
travelr 0.7.5 -> 0.9.1
tzdb 0.1.2 -> 0.2.0
usethis 2.0.1 -> 2.1.3
xfun 0.24 -> 0.27
Cool hey?
A lot of the automations I rig up in my code editor depend on decting where the cursor is in a document and using that context to perform helpful operations.
The simplest class of these are functions that are executed using the symbol the cursor is “on” as input. Typically this symbol represents an object name and typical usage would be:
str() on the object to inspect ittargets::tar_load() on the object to read it from cache into the global environmentSimple things that help keep my hands on the keyboard and my head in the flow.
RStudio poses two challenges in setting these types of things up as keyboard shortcuts:
To solve 1. we can use Garrick Aden-Buie’s {shrtcts} package. To solve 2. there’s a tiny package I wrote called {atcursor}.
Suppose we desire a shortcut to call head() on the object cursor is on. This is how we could rig that up in ~/.shrtcts.R:
#' head() on cursor object
#'
#' head(symbol or selection)
#'
#' @interactive
function() {
target_object <- atcursor::get_word_or_selection()
eval(parse(text = paste0("head(",target_object,")")))
}
After that we’d:
shrtcts::add_rstudio_shortcuts(){shrtcts} can also manage the keyboard bindings with an @shortcut tag but add_rstudio_shortcuts() won’t refer to it by default. See the doco if you want to do that.atcursor::get_word_or_selection() will return a symbol the cursor is “insisde” - e.g. on a column inside the span of the string. If the symbol is namespaced the namespace is also returned, e.g: “namespace::symbol”. If the user has made a selection, that is returned, regardless of cursor position.Rather than building text to parse and then eval, sometimes I find it easier to work with expressions. So you coud do like: target_object <- as.symbol(atcursor::get_word_or_selection()) and then build an expression with bquote:
eval(bquote(
some(complicated(thing(.(target_object))
))
Without getting overly metaphysical: I think these kind of shortcuts make a lot of sense to me because I view the cursor as my avatar in this world of code before me. I navigate that world almost exclusively with keys, so coding is like piloting that little avatar around. To learn about objects or manipulate them, it makes complete sense to cruise up to them and start engaging them in a dialogue of commands, the scope of which is completely unambigous, because my avatar is in the same space as those objects. In this way, my sense of ‘where I am’ in the code is not broken.
Ofcourse it does happen, I have to jump to the console world when I don’t have a binding for what I need to do, but it feels great when I don’t!
If anyone else is down for some command line JSON munging this little tool knocked my socks of this week: stedolan.github.io/jq/